|
I like finding new ways to present data and someone posted about this Ancestor Spiral today (link Here) on the Genealogy Addicts Anonymous Facebook Group. You can use any long string of words, but when an ancestor list is used, it resembles the rings of a tree trunk, with the older ancestors in the center and the more recent ones on the outer rings.
4 Comments
The formatting of the blog posting may be odd if you are reading this in a Feed Reader or via e-mail distribution, so click on the title above (which is an active link) to view the website version.
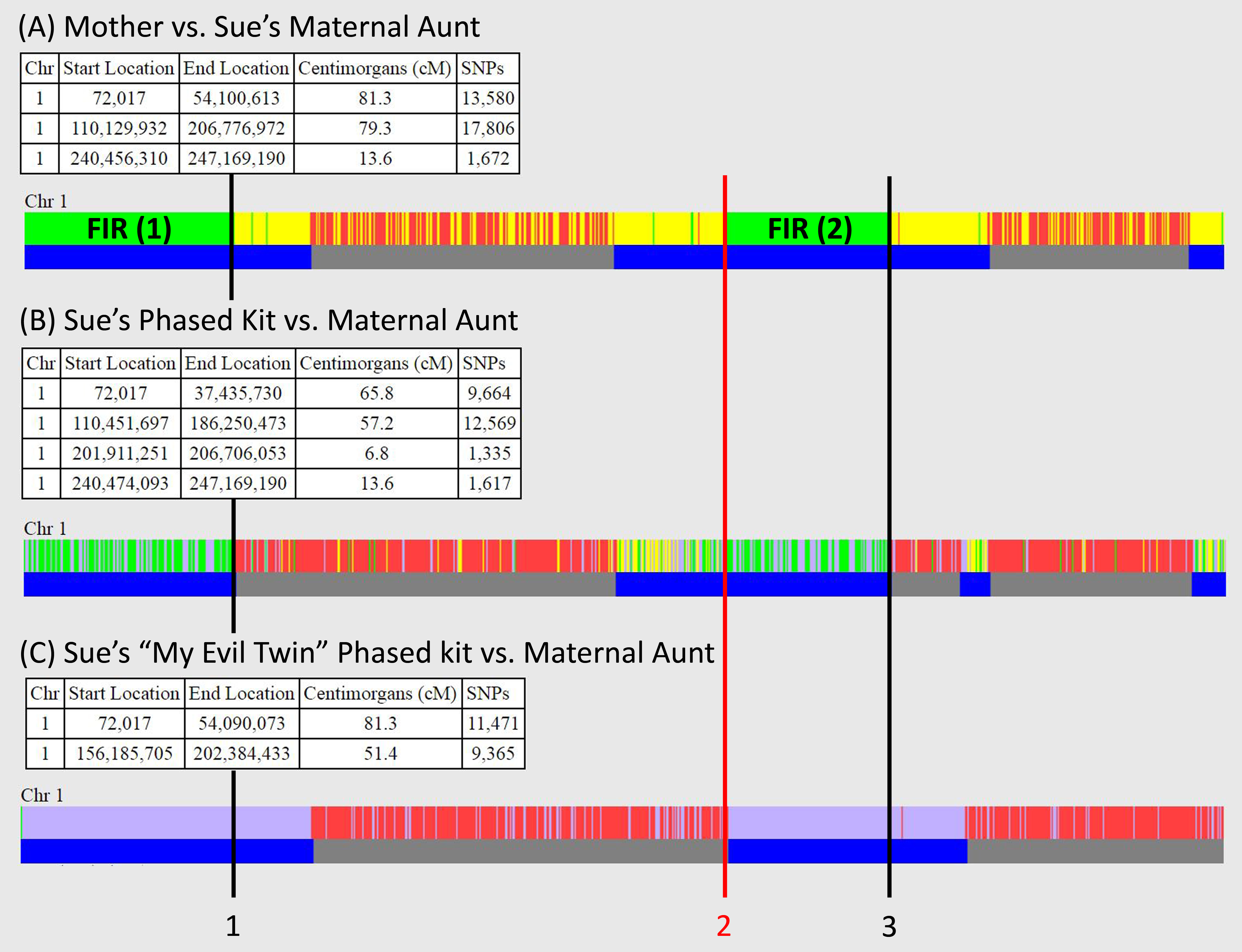
Since my blog posting last month, Obtaining FIR Boundaries on GEDmatch using the Little Tick Marks, GEDmatch has introduced a new Tier 1 tool, known as "My Evil Twin" Phasing, which makes it much easier to obtain the boundaries for FIRs (fully-identical regions) if at least one child's results are available; this is used in combination with the normal phasing tool.
Figure 1 shows one-to-one comparisons on GEDmatch for Chromosome 1:
The formatting of the blog posting may be odd if you are reading this in a Feed Reader or via e-mail distribution, so click on the title above (which is an active link) to view the website version.
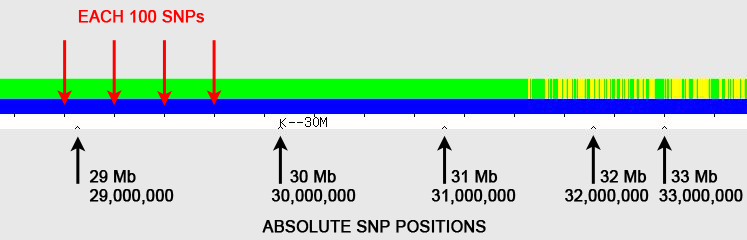
Question: How do you obtain the boundaries of FIRs (fully-identical regions) on GEDmatch?
Answer: Using the little "inverted V" tick marks on the one-to-one tool graphic (see Figure 1). The red arrows point to regularly spaced tick marks, which identify every 100 SNPs (single nucleotide polymorphisms). The black arrows point to irregularly spaced "inverted V" tick marks; these show the absolute SNP positions and are spaced at 1 Mb (1,000,000) intervals, with labels every 10 Mb (10,000,000) – in the example here, the label "K--30M" represents SNP position 30 Mb (30,000,000). [Someone has pointed out that the "K" on GEDmatch is actually a vertical line with left-pointing arrow (<--), which I agree is almost certainly the case, but I've continued to use "K" in this blog posting as it is simplistically descriptive.] Follow-up Question: But where are the little tick marks?
The formatting of the blog posting may be odd if you are reading this in a Feed Reader or via e-mail distribution, so click on the title above (which is an active link) to view the website version.
Elizabeth "Bessie" Thomas (1851-1930) was brought up in Llanerchymedd (pronunciation) on the island of Anglesey in Wales. In 1869, when she was 18 years old, she wrote the following poem in the front cover of her bible. The last line of her poem is particularly poignant, as she is now being remembered on the internet and her words potentially seen by far more people than she could have ever envisioned.
The formatting of the blog posting may be odd if you are reading this in a Feed Reader or via e-mail distribution, so click on the title above (which is an active link) to view the website version.
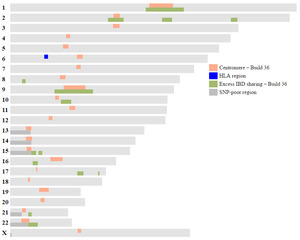
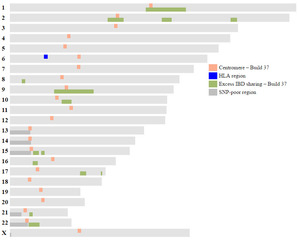
I do a lot of chromosome mapping using Kitty Cooper's great Ancestor Mapping Tool. There are a few regions in the genome more likely to have issues when comparing matching DNA segments with others: at the centromeres and regions of excess IBD sharing (IBD = identical by descent). Instead of looking these up in tables, I've plotted them on chromosome maps, as visual representations are easier than looking up values in a table and they allow side-by-side comparisons to be made to my chromosome maps. I also added the Human Leukocyte Antigen (HLA) region on chromosome 6, which exhibits greater IBD sharing than expected, and the SNP-poor regions (SNP = single nucleotide polypeptide) rather than having blank areas on the chromosome map.
 DNArboretum DNArboretum
Thanks to a posting on the Yahoo! DNA-Newbie message board (Topic 49775), I'm now aware of a great FREE extension for Google Chrome, which was released in April 2014 by "neanderling" (if you have a 23andMe account, see thread Here). I hadn't seen it mentioned elsewhere, so thought a blog posting would spread the word, as I'm certain that others will find it useful too.
Let's face it, the trees at 23andMe and Family Tree DNA (FTDNA) are pretty awkward and clunky. DNArboretum quickly extracts information from a displayed tree and presents it as a list based on Ahnentafel number, showing the name and, if included on the tree, the year and location of birth and death. You can then either skim down the list, or search for names or locations. This is so much easier than trying to wade through the trees on either 23andMe or FTDNA. Unfortunately, it only works on the old-style trees on 23andMe – for anyone with a tree that is now on MyHeritage, the only way a tree can now be set up (see 23andMe and MyHeritage Collaboration), DNArboretum doesn't work, but there are still some old-style trees on 23andMe if people didn't "upgrade" to a tree on MyHeritage. When available, at least 9 generations are displayed at 23andMe and up to 15 generations at FTDNA.
The other nice feature of DNArboretum is that you have the option to show different inheritance modes: all ancestors (autosomal mode), or only those on the X-chromosome DNA, mitochondrial DNA, or Y-chromosome DNA lines.
The formatting of the blog posting may be odd if you are reading this in a Feed Reader or via e-mail distribution, so click on the title above (which is an active link) to view the website version.
The formatting of the blog posting may be odd if you are reading this in a Feed Reader or via e-mail distribution, so click on the title above (which is an active link) to view the website version.
The formatting of the blog posting may be odd if you are reading this in a Feed Reader or via e-mail distribution, so click on the title above (which is an active link) to view the website version.
I was aware that AncestryDNA would be reducing the number of DNA matches to something more reasonable (see, for example, When Less is More), so I was fully prepared for a significant reduction in DNA matches for myself and my parents. However, I expected some consistency with what would be done (whether I agreed with it or not), but in my limited experience, this is not what has happened.
The formatting of the blog posting may be odd if you are reading this in a Feed Reader or via e-mail distribution, so click on the title above (which is an active link) to view the website version.
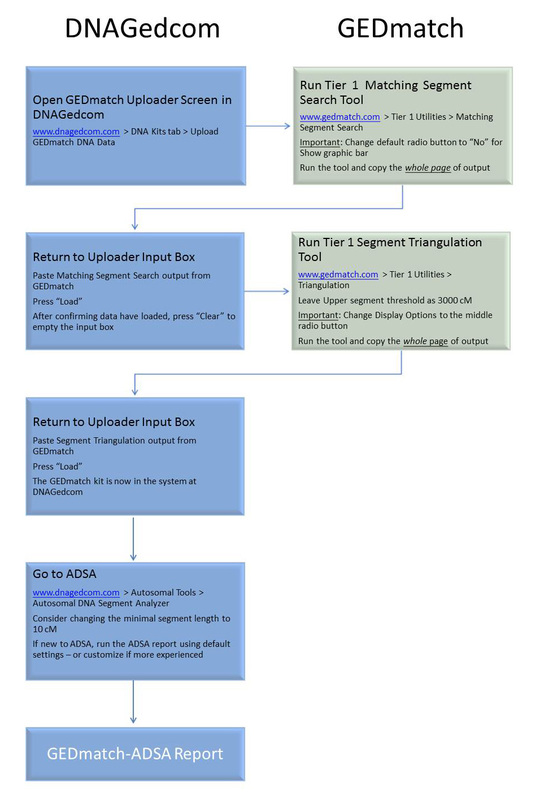
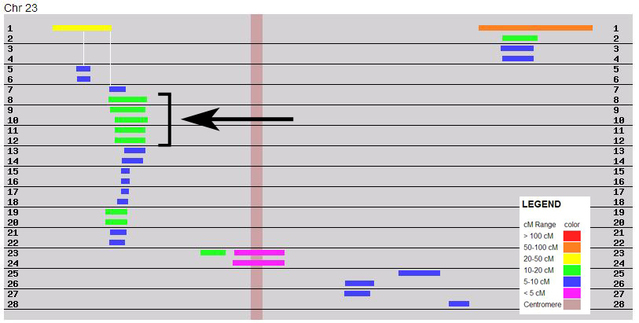
Those of us who are heavy users of GEDmatch are sorely missing the wonderful segment triangulation tool, which hasn't yet been restored since the relocation of GEDmatch to new servers. In order to be able to continue identifying triangulated groups with DNA matches, I'm using a workaround, by expanding what I previously did for the X-chromosome (Chr 23 in the figure below), which isn't included in the segment triangulation tool. In addition, segment triangulation on GEDmatch is limited to the highest 200 matches, so this method can also be used for lower level matches, even when the segment triangulation tool is back.
|
AuthorSee About Me
Contact MeSubscribe to Blog
Blog CategoriesArchives
August 2019
|