The formatting of the blog posting may be odd if you are reading this in a Feed Reader or via e-mail distribution, so click on the title above (which is an active link) to view the website version.
Question: How do you obtain the boundaries of FIRs (fully-identical regions) on GEDmatch?
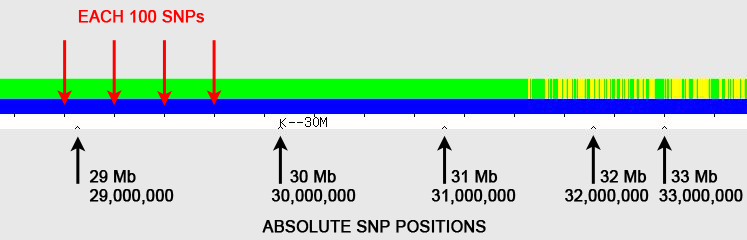
Answer: Using the little "inverted V" tick marks on the one-to-one tool graphic (see Figure 1).
The red arrows point to regularly spaced tick marks, which identify every 100 SNPs (single nucleotide polymorphisms). The black arrows point to irregularly spaced "inverted V" tick marks; these show the absolute SNP positions and are spaced at 1 Mb (1,000,000) intervals, with labels every 10 Mb (10,000,000) – in the example here, the label "K--30M" represents SNP position 30 Mb (30,000,000). [Someone has pointed out that the "K" on GEDmatch is actually a vertical line with left-pointing arrow (<--), which I agree is almost certainly the case, but I've continued to use "K" in this blog posting as it is simplistically descriptive.]
Follow-up Question: But where are the little tick marks?
Answer: Using the little "inverted V" tick marks on the one-to-one tool graphic (see Figure 1).
The red arrows point to regularly spaced tick marks, which identify every 100 SNPs (single nucleotide polymorphisms). The black arrows point to irregularly spaced "inverted V" tick marks; these show the absolute SNP positions and are spaced at 1 Mb (1,000,000) intervals, with labels every 10 Mb (10,000,000) – in the example here, the label "K--30M" represents SNP position 30 Mb (30,000,000). [Someone has pointed out that the "K" on GEDmatch is actually a vertical line with left-pointing arrow (<--), which I agree is almost certainly the case, but I've continued to use "K" in this blog posting as it is simplistically descriptive.]
Follow-up Question: But where are the little tick marks?