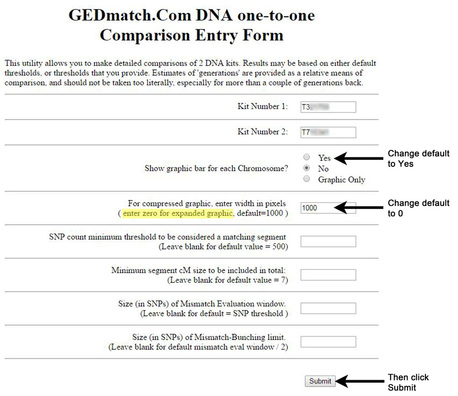
Answer: Using the little "inverted V" tick marks on the one-to-one tool graphic (see Figure 1).
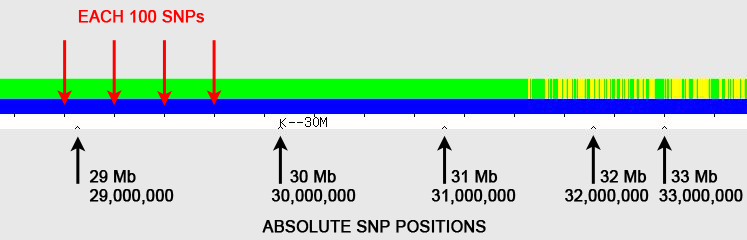
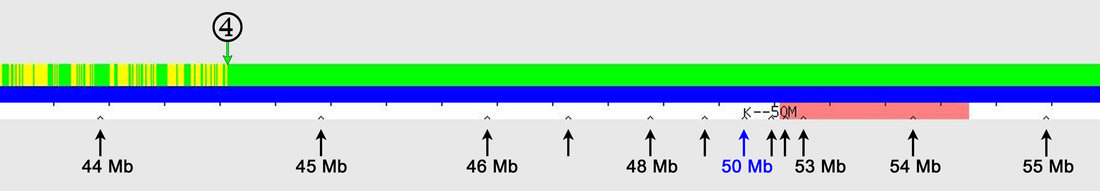
The red arrows point to regularly spaced tick marks, which identify every 100 SNPs (single nucleotide polymorphisms). The black arrows point to irregularly spaced "inverted V" tick marks; these show the absolute SNP positions and are spaced at 1 Mb (1,000,000) intervals, with labels every 10 Mb (10,000,000) – in the example here, the label "K--30M" represents SNP position 30 Mb (30,000,000). [Someone has pointed out that the "K" on GEDmatch is actually a vertical line with left-pointing arrow (<--), which I agree is almost certainly the case, but I've continued to use "K" in this blog posting as it is simplistically descriptive.]
Follow-up Question: But where are the little tick marks?
First a few abbreviations, definitions, and explanations:
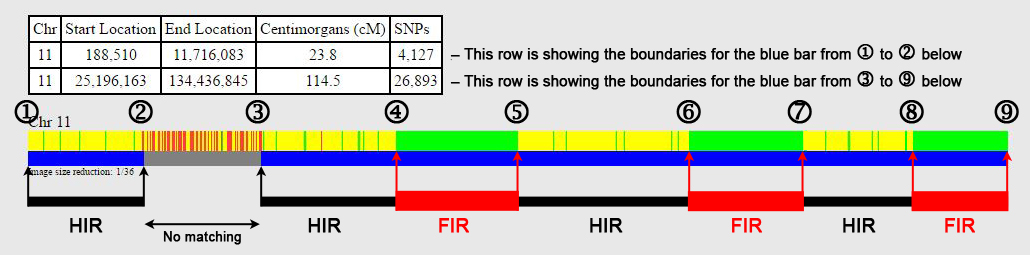
HIR: Half-identical region (see Here for information on the ISOGG wiki), where the alleles are matching along a stretch of DNA on either the maternal or paternal chromosome – this is the typical type of matching seen.
FIR: Fully-identical region (see Here for information on the ISOGG wiki), where the alleles are matching along a stretch of DNA on both the maternal and paternal chromosomes – seen when comparing full siblings and double 1st cousins (who also have HIRs).
SNP: Single nucleotide polymorphism (see Here for information on the ISOGG wiki)
David Pike has a number of free DNA tools, including Search for Shared DNA Segments in Two Raw Data Files, which will give the start and end locations for both HIRs and FIRs, but you need to have access to the raw data files from both individuals to run this. However, by using the "inverted V" tickmarks on GEDmatch, it is quick to make a pretty good estimate of the start and end positions of the FIRs, although you have to check each one individually.
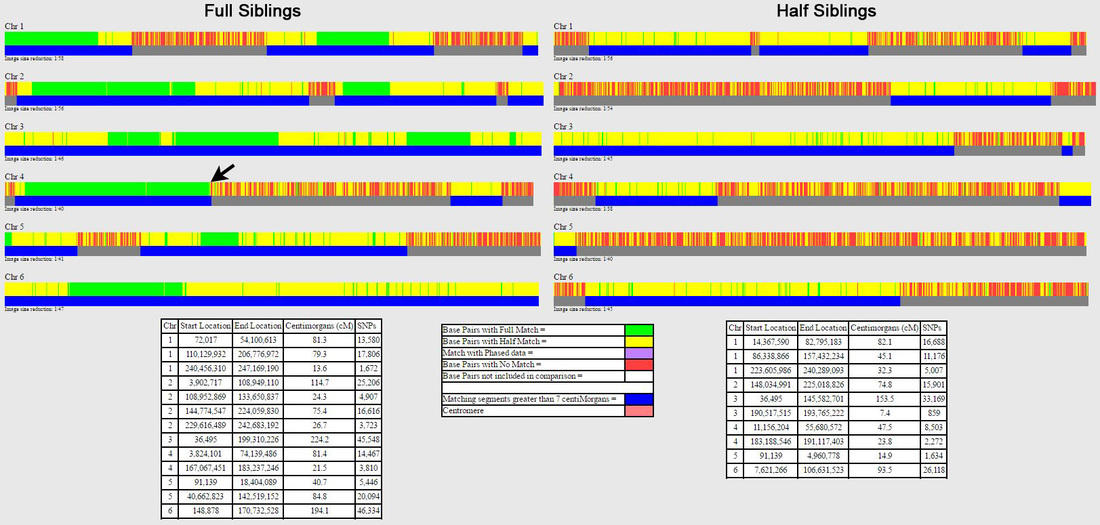
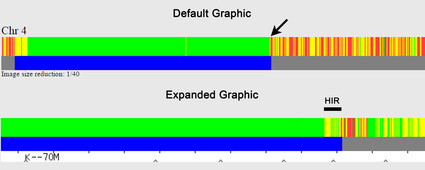
The expanded view is also useful for evaluating the position of (almost) coincidental hidden crossovers in siblings, where a matching segment is far too short to be shown as matching (on either GEDmatch or using David Pike's tool – not shown here), or where the matching segment appears to go directly from a FIR to a non-matching segment (without an intervening HIR), which is a different example of an (almost) coincidental crossover. The latter is shown by the black arrow for full siblings in Figure 2, which has been enlarged in the top panel of Figure 6 – the lower panel shows the expanded view, where it can be seen that there is actually a short HIR between the green FIR and the non-matching segment, which isn't apparent on the default one-to-one graphic. The one-to-one comparisons in Figure 6 are for my mother and her sister (her full sibling), and because I've done a lot of chromosome mapping, I've worked out that my aunt inherited a paternal crossover at ~73.8 Mb and a maternal crossover at ~74.1 Mb, which are almost coincidental.
Lastly, although most of the tools on GEDmatch are free, don't forget that it is not a commercial enterprise and relies on donations for server and other costs. Please consider a donation via Paypal – there is a link at the bottom of your Home page. Donating helps all of us, as it enables GEDmatch to enhance what they can offer.